Authors:
- Nurudin Alvarez-Gonzalez
- Andreas Kaltenbrunner
- Vicenç Gómez
tl;dr
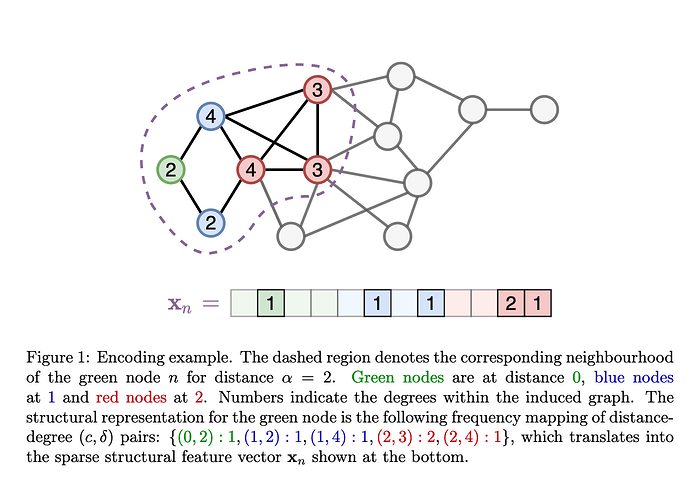
Creates features of a node by encoding the degree and distance of the nodes in the neighborhood. Neighborhood size is a hyperparameter, \alpha. The size of the resulting vector is \alpha \cdot d_\text{max}. The values of this (sparse) vector are the count of the number of nodes that satisfy that degree/distance criteria.
This local encoding is then simply passed through a linear layer to densify and embed.
Experimental Setting
Datasets
- Les Miserables
- Sister Cities
- ArXiv AstroPhysics
- PPI
Baselines
- Unattributed graph embeddings (all transductive):
- DeepWalk
- node2vec
- LINE (Line: Large-scale information network embedding)
- VERSE (Verse: Versatile graph embeddings from similarity measures)
- Inductive methods (all require node attributes):
- GCN
- GraphSAGE
- LGCL (large-scale learnable graph convolutional networks)
- PinSAGE
Evaluation metrics
Unsupervised
Samples random walks from each node. Then use a skip-gram negative sampling objective.
Run this to generate embeddings and then cluster on the embeddings. In Les Miserables (which have two copies of the graph with a single edge between a random pairing across graphs), they find that by setting the receptive field of the structure encoding to \alpha=1, clusters mostly capture densely connected nodes. However, when \alpha=2, cluster assignment is more based on structural role and less so on node proximity.
Also look at ROC-AUC on Link Prediction tasks using Facebook and ArXiv datasets.
Supervised
Exactly what you’d expect–use these as input embeddings to a downstream model.
Multi-label node classification task where the objective is to predict 121 different binary labels capturing protein functions in a series of Protein-to-Protein Interaction (PPI) graphs.