This post is an experiment in doing open research. It’s unclear what general value it will offer, if any. The plan will be to document my thinking and progress as they develop. Comments/questions are encouraged.
Motivation
In practice, many real-world graphs of interest evolve in time. For example, graphs representing users and their connections for purposes of fraud detection, or their interactions with products for recommendation systems, both have a constant stream of new nodes and edges, and potentially dynamic features that change in time. This presents at least 4 distinct problems for traditional GNNs:
- The majority of GNN methods in the literature are transductive, meaning they do not generalize to unseen nodes/edges. This is an egregious problem because it’s often the new items that are of particular interest and in need of a prediction. In practice, these methods need to be retrained when the graph changes, and this presents a number of operational problems. We therefore need an inductive model to be useful in most practical applications.
- Most GNN methods assume a static graph and have no capability to leverage temporal information. Temporal information is key in many problems, however. For example, a rapidly emerging cluster of users is perhaps more suspicious in fraud applications, or predictive of a viral/timely piece of content in a recommendation context.
- Popular GNN modeling frameworks, like Deep Graph Library and PyTorch Geometric, were not designed to elegantly handle time-evolving graph data structures, features, or time-aware message-passing operations. As a result, implementing time-aware models is often difficult, inefficient, or reliant on approximation schemes, like treating the graph as a series of static snapshots that trade-off memory with approximation quality. Simple mechanisms like edge masking can be employed for dynamic edges, but handling evolving features presents additional complications.
- The static graph assumption (and the frameworks built on this) causes yet another problem: message passing operations themselves are not time-aware and as a result, models often leak information from the future during training. The models do not have this advantage in a production setting, of course, and therefore suffer in performance.
- As an example, in the
ogbn-arxivbenchmarks, the graph methods first reverse the edges, meaning that a classification decision for a paper not only depends on what it cites, but also on who cites it in the future. If the goal is to classify new papers, clearly these future citations will not be available and this model’s real-world performance will be far lower than what’s reported on the test set.
- As an example, in the
In sum, there’s a significant disconnect between existing GNN methods, frameworks and benchmarks, and many real-world industrial problems. As a consequence, many approximations and compromises need to be made.
Hypothesis
My hypothesis is that by sampling a subgraph around a target node, we can use transformers to aggregate this information into a prediction that more naturally models the problem, particularly for dynamic graphs. Instead of having a large graph structure that’s used for message passing, we instead pre-process our data to construct a collection of subgraphs. These choices give us a number of benefits:
- The model is trained on localized subgraphs and is therefore naturally inductive, and will generalize to new subgraphs.
- Temporal information can be naturally represented via temporal encodings of the entities, allowing the model to leverage temporal dynamics without significant incremental effort.
- There exists a large community investment in transformer tooling and this method removes the need to rely on the complexities of GNN training and frameworks, like handling message passing in distributed training environments. This approach can be trivially parallelized.
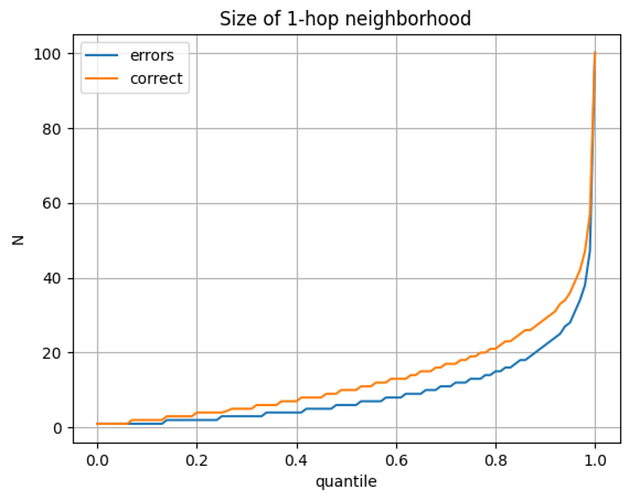
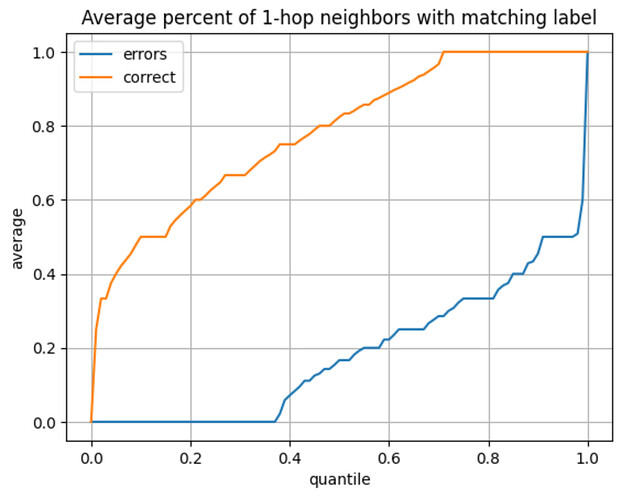
- Since we do not need a single representation of connection structure, each neighborhood snapshot can be point-in-time correct relative to the target node. This removes the need to have in-memory data structures that can elegantly handle all the time-evolving information and prevents future information leakage. Unlike in GNNs, this allows us to, for example, easily include neighbor labels as features.
- GNNs need to resort to various tricks to handle this use case of “labels as features”, like stochastically removing label features during training. See the Label Usage section here for an example.
Challenges
The key challenge here is figuring out how to usefully encode graph structure so that the transformer can make efficient use of it. There are a few ideas:
- Use an approach similar to Pure Transformers are Powerful Graph Learners, which encodes a graph structure by concatenating positional representations of the source/destination pairs of nodes that have edges: \{ P_u \Vert P_v : (u, v) \in \mathcal E \}, where the P_i positional encodings are just required to be orthonormal.
- Learn graph-aware node encodings based on things like:
- Shortest path length from the target node
- Node type, or shortest meta-path to target node, for heterogeneous graphs
- A structural property of the nodes, like degree, or “color” resulting from running the W-L algorithm
- Some combination of the above